Current research activities
Riboflavin (vitamin B2) is a universal precursor for flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) which are essential cofactors for many flavoenzymes that carry out great diversity of redox reactions. These flavoenzymes participate in a wide range of biological processes, including carbohydrate, amino acid, and fatty acid metabolisms. While yeast and other higher animals obtain riboflavin from their diet, bacteria and some plants synthesize these cofactors by de-novo method. In bacteria, seven enzymes (ribA, ribB, ribC, ribD, ribE, ribF and ribH) are involved in the biosynthesis of these cofactors (riboflavin, FMN and FAD) using GTP and ribulose-5-phosphate as initial substrates. Since these enzymes are shown to be essential for the bacterial growth and not present in human, they are considered as potential targets for the development of anti-bacterial drugs. Our lab is interested in establishing the structure-function relationship of these bacterial enzymes by X-ray crystallography. The three-dimensional structure available from these studies will provide a platform for rational structure based drug design to identify potential lead molecules which can kill pathogenic bacteria specifically. Some of the representative structures from riboflavin biosynthesis pathway is shown here.
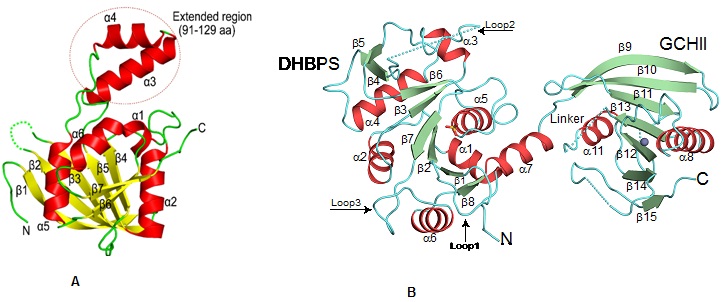
Cartoon representation showing the overall fold of (A) ribA (GCHII) enzyme from Helicobacter pylori (Journal of Structural Biology, 2015, 192, 100-115).(B) Bifunctional ribA (GCHII) and ribB (DHPBS) enzyme from Mycobacterium tuberculosis(ActaCryst, 2013, D69, 1633-1644).
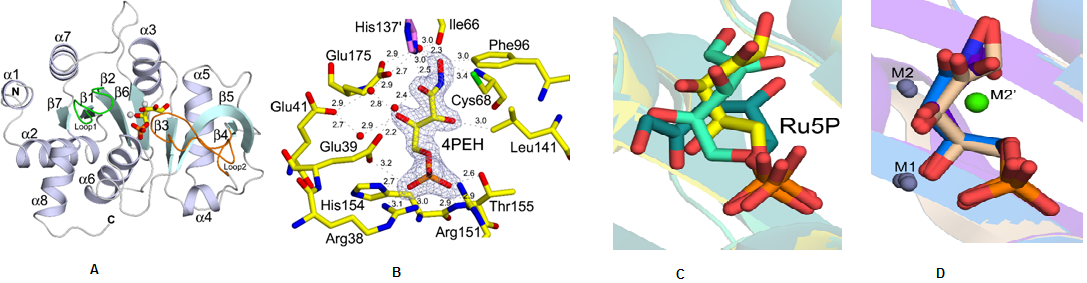
Structural characterization of ribB from Vibrio cholerae. (A) Cartoon representation showing the overall fold of ribB (DHBPS)(B) Showing a 2Fo-Fc electron densitymapcontoured at 1.2sfor along with an inhibitor 4PEH. (C)Superposition of substrate Ru5Pin the absence of metal ions from different species(D) Superposition of substrate Ru5P and inhibitor 4PEH in thepresence of metal ions (Journal of Biological Chemistry, 2015, 290, 11293-11308).
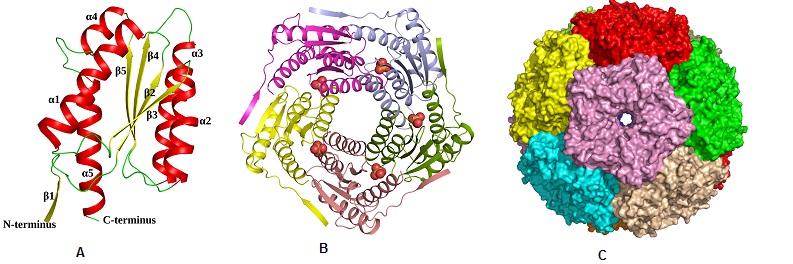
Structural characterization of ribH from Salmonella typhimurium. (A) Cartoon representation showing the overall fold of ribH (Lumazine Synthase) (B) Pentamer assembly of lumazine synthase; each subunit is represented in different colour. (C)Surface representation of the icosahedral assembly of lumazine synthase, showing each pentamer in a different colour (ActaCryst, 2011, D67, 131-139)
A key focus of our laboratory is on studying new types of "molecular recognition" mechanism(s) with an aim to establish a novel framework for the design of intelligent drugs which will exhibit more selectivity and potency towards their targets, as compared to their conventional counter parts, orthosteric and allosteric drugs. Our approach involves a combination of structural, computational, and analytical methods, with hypothetical models to test and validate yet undiscovered molecular recognition principles. By using above approaches, we recently discovered a competitive-allosteric mechanism by which a low-affinity ligand can displace a high-affinity lignd bound to the target by not only compete for the same site but also by inducing specific allsoteric structural changes to the target which accelerates the dissociation of the high-affinity competitor. Currently, our efforts towards establishing a experimental framework for rapid identification of "competitive-alosteric" ligands and validating their relevance for pharmaceutical industry.
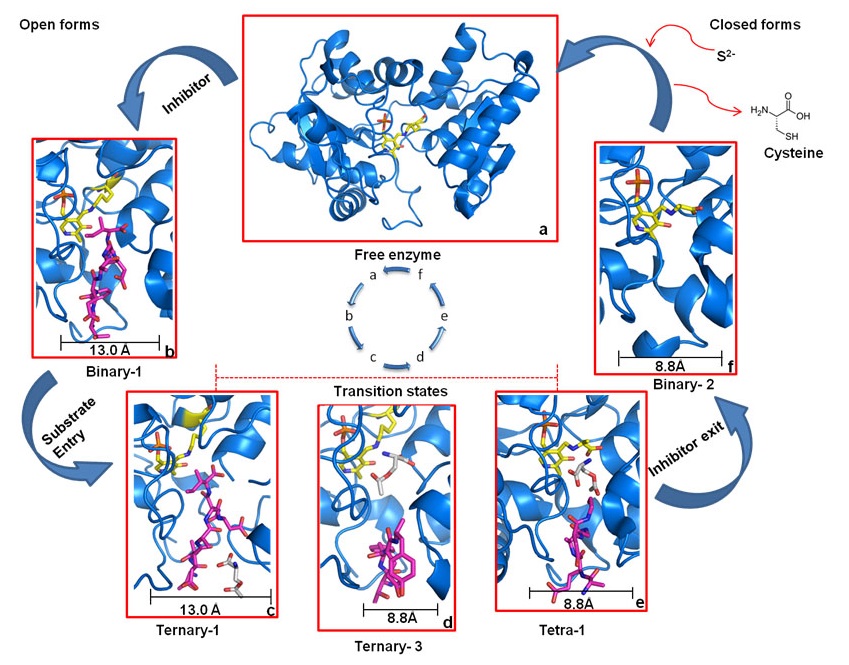
Dissociative cycle of enzyme-inhibitor complex
High-affinity inhibitor is shown as magenta sticks and enzyme, marine blue. a) Free enzyme in the open-state, ready to bind inhibitor. b) OASS·inhibitor complex in open-state. c) Binding of low-affinity substrate to enzyme·inhibitor complex. d) Translocation of substrate to reaction center and dissociation of inhibitor from S1 site. e) Reaction of the substrate, and exit of the inhibitor. f) Inhibitor is expelled and OASS is trapped in enzyme·reaction-intermediate complex in the final closed state, ready to react with incoming sulfide to form cysteine. After cysteine release, OASS switches back to open-state (a) to start the cycle again