Data set
B-cell epitopes were obtained from B cell epitope database (
BCIPEP), which contains 2479 continuous epitopes, including 654 immunodominant, 1617 immunogenic epitopes. All the identical epitopes and non-immunogenic peptides were removed, finally we got 700 unique experimentally proved continuous B cell epitopes. The dataset covers a wide range of pathogenic group like virus, bacteria, protozoa and fungi. Final dataset consists of 700 B-cell epitopes and 700 non-epitopes or random peptides (equal length and same frequency generated from
SWISS-PROT).
Neural Networks
An artificial neural network (ANN) is an information-processing paradigm inspired by the way the densely interconnected, parallel structure of the mammalian brain process information. Artificial neural networks are collections of mathematical models that emulate some of the observed properties of biological nervous system and draw on the analogies of adaptive biological learning. The key element of the ANN paradigm is the novel structure of the information processing system. It is composed of a large number of highly interconnected processing elements that are analogous to neurons and are tied together with weighted connections that are analogous to synapses.
The ABCpred server uses partial recurrent neural network (Jordan network) with a single hidden layer. The networks have optional window length of 10, 12, 14, 16, 18 and 20, and have 35 residues in a single hidden layer. The target output consists of a single binary number and is 1 or 0 (epitope or non epitope). For neural network implementation and to generate the neural network architecture and the learning process, the publicly available free simulation package SNNS4.2v from Stuttgart University (Zell and Mamier, 1997) is used. It allows incorporation of the resulting networks into an ANSI C function for use in stand-alone code.
Working with Partial recurrent Networks
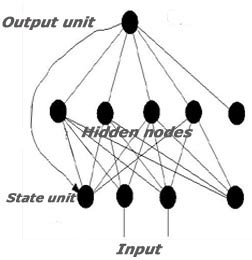 Recurrent neural network
|
In this section, the initialization, learning, and update functions for partial recurrent networks are described.
(i)
The initialization function JE_Weights
The initialization function JE_Weights requires the specification of five parameters.
- alpha, beta: The weights of the forward connections are randomly chosen from the interval [alpha;Beta];
- lamda: Weights of self recurrent links from content units to themselves. The network use lamda=0;
- gamma: Weights of the other recurrent links to content units. This value is set to 1.0;
- phi: Initial activation of all context units.
(ii)
Learning functions
The total network input consists of two components. The first component is the pattern vectors, which was the only input to the partial recurrent network. The second component is a state vector. This state vector is given through the next-state function in every step. By this way the behavior of a partial recurrent network can be simulated with a simple feedforwaded network, that receives the state not implicitly through recurrent links but as an explicit part of the input vector. In this sense, the backpropagation algorithm can easily be modified for the training of partial recurrent networks.
The learning function has been adapted for the training of partial recurrent Jordan networks.
JE_BP standard Backpropagation has been used for this partial recurrent network.
(iii)
Update functions
The update function which has been implemented for partial recurrent network is JE_Order. This update function propagates a pattern from the input layer to the first hidden layer, then to the second hidden layer, etc. and finally to the output layer. After this follows a synchronous update of all context units.
Measure of prediction accuracy
Both threshold dependent and independent measures have been used to evaluate the prediction performance.
Threshold dependent measures
Three parameters were used to measure the performance of prediction methods. Following is the brief description of these parameters:
(i)
Sensitivity (Qsens) of the prediction methods is defined as the percentage of epitopes that are correctly predicted as epitopes. Higher sensitivity means that almost all of the potential epitopes will be included in the predicted results.
Qsens=(TP/TP+FN)*100
Where TP and FN refer to true positives and false negatives respectively.
(ii)
Specificity (Qspec) of the prediction methods is defined as the percentage of correctly predicted non-epitopes.
Qspec=(TN/TN+FP)*100
Where TN and FP refer to true negatives and false positives.
(iii) A commonly used parameter to measure the prediction performance is
accuracy, is defined as
Qacc=(TOTc/Total)*100
TOTc is the total number of correct predictions (includes both true positives, TP and true negatives, TN) and Total is the total number of prediction made.
(iv)
Positive prediction Value(Qppv) measures the probability that a predicted epitopes is in fact a epitope.
Qppv=(TP/TP+FP)*100
Threshold independent measures-ROC
One problem with the threshold dependent measures is that they measure the performance at a given threshold. They fail to use all the information provided by a method for evaluation. The Receiver Operating Characteristics (ROC) is a threshold independent measure that has been developed as a signal processing technique (Deleo, 1993). A ROC plot is obtained by plotting all sensitivity values (true positive fraction) on the y-axis against their equivalent (1 - specificity) values (false positive fraction) for all available thresholds on the x-axis. The area under the ROC curve measures discrimination, the ability of a method to correctly classify epitopes and non-epitopes.
|







