Help Page
We designed our web-server VitaPred in very user friendly mode. Where any user can submit their protein sequence for the prediction of vitamin-interacting residues. Here we describing the way how to use VitaPred easily. The Job submission page includes following options and sections, which you can use according to your need.
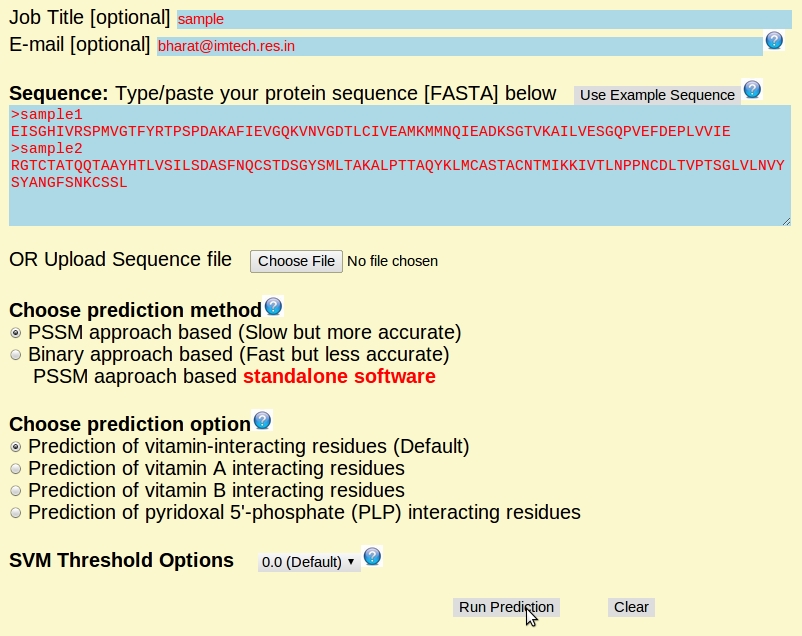
Job Title [optional]: User can give a specific name for job title.
E-mail [optional]: All prediction results will also send to the given Email ID.
Sequence: This web-server allows only one protein sequence at a time for the prediction of vitamin interacting residues. The protein sequence should be in standard FASTA format. User can also upload their protein sequence.
Choose prediction method: We have provided three different method - (1) PSSM approach based (Slow but more accurate), (2) Binary approach based (Fast but less accurate), and (3) PSSM aaproach based standalone software. User can use any method according to thier need.
Choose prediction option: We have provided four different options: (1) Prediction of vitamin-interacting residues (Default); (2) Prediction of vitamin A interacting residues; (3) Prediction of vitamin B interacting residues; and (4) Prediction of pyridoxal 5'-phosphate (an active form of vitamin B6) interacting residues.
SVM Threshold Options: SVM performances is threshold-dependent, so please choose appropriate threshold level as per your requirement. The probability of correct prediction directly depends on the threshold. The default threshold of VitaPred is -0.8. If user wants less sensitivity but more specificity, then higher threshold value should be specified, if the requirement of user is more sensitivity and less specificity then lower threshold value should be choosen. So, the expected outcome will depends on the trade-off between sensitivity and specificity.
The overview of submission page: (an example)
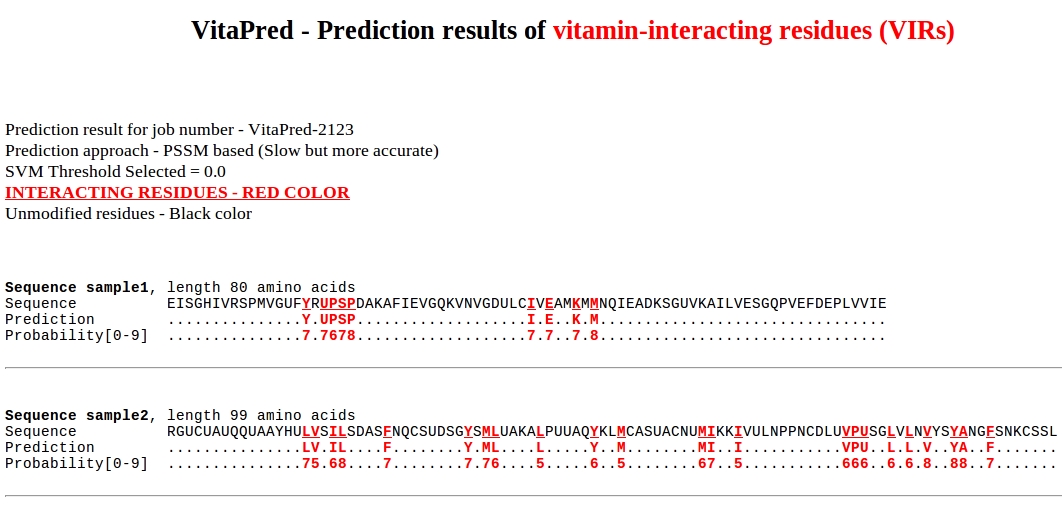
The overview of prediction result page: (an example)