Algorithm of DNAint
Support Vector Machine (SVM)
SVM is a well established tool for the biological predictions. We applied and optimized different parameters/kernels of SVM for the development of DNAint.
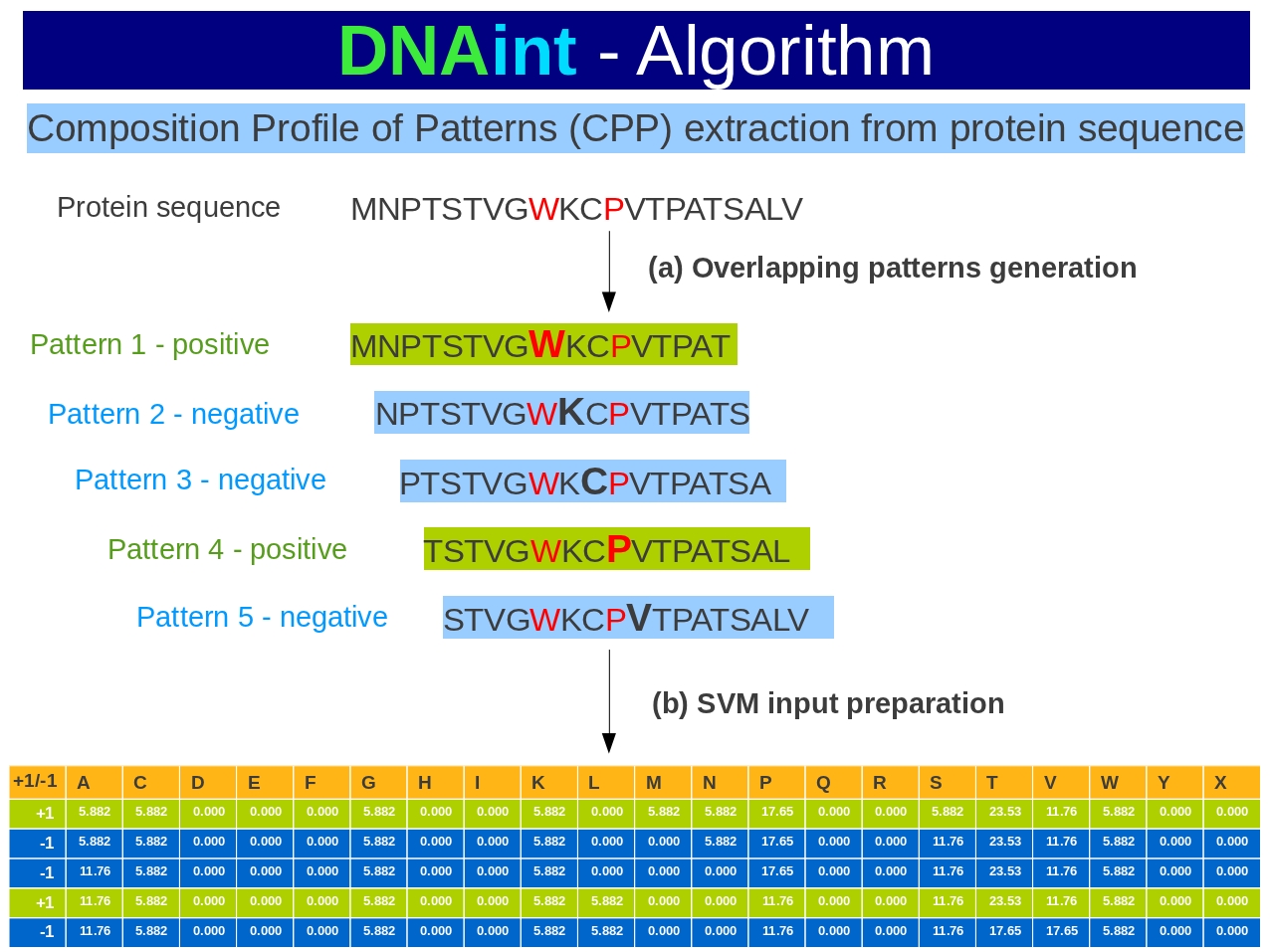
Composition Profile of Patterns (CPP)
The overlapping patterns were generated from protein sequences and calculated the amino acids compositions of all patterns separately.These compositions termed as Composition Profile of Patterns (CPPs) and used for the development of SVM based DNA-interacting residues (DIRs) prediction. The following diagram showing overview of DNAint algorithm.
Two sample logo of DIRs and non-DIRs
Two Sample Logo (TSL) of DNA interacting as positive and non-interacting as negative patterns of 17-length. The 9th position residue is either DIR or non-DIR in the pattern.
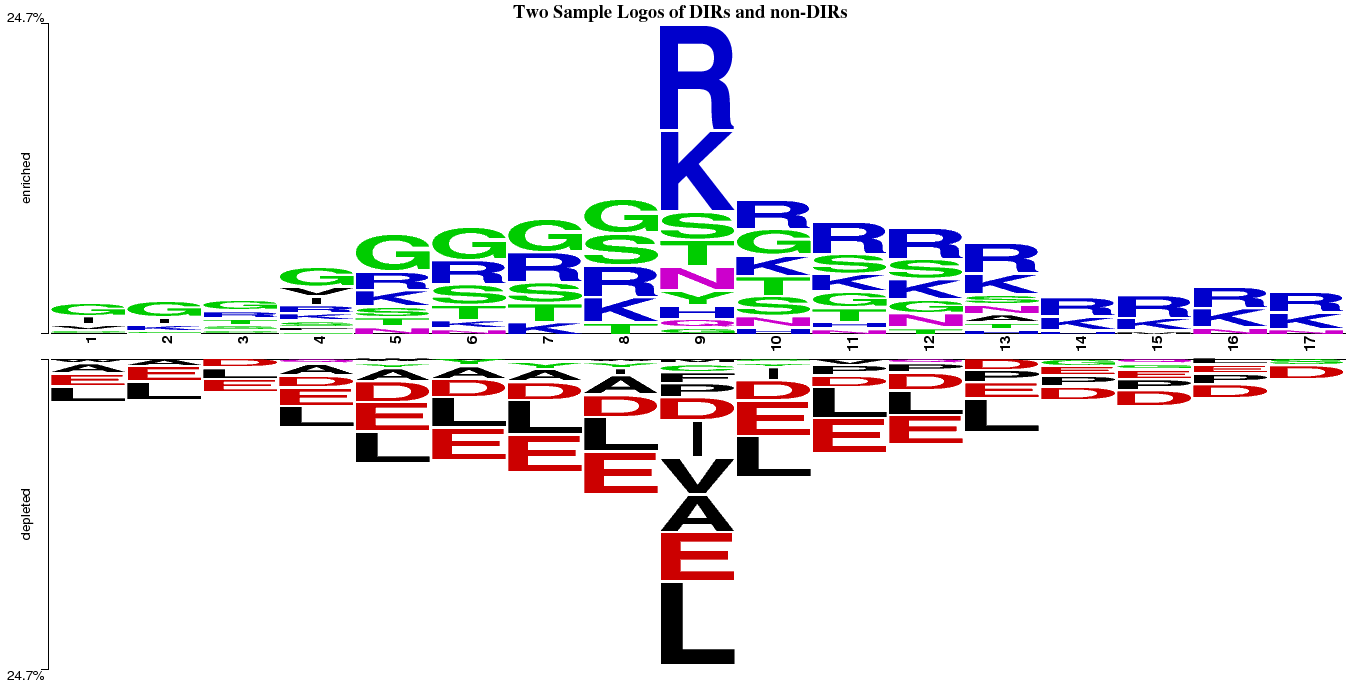 Two sample logo of DNA interacting and non-interacting patterns
Two sample logo of DNA interacting and non-interacting patternsEvaluation Methods
The prediction models were evaluated by five-fold cross validation techniques using the following formulas :-
Sensitivity = (TP / (TP+FN))*100
Specificity = (TN / (TN+FP))*100
Accuracy = (TP+TN / (TP+FP+TN+FN))*100
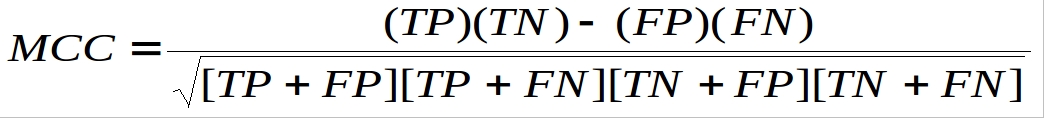
Where TP and TN are correctly predicted DIRs and non-DIRs respectively. FP and FN are wrongly predicted DIRs and non-DIRs respectively.
Probability Score
DNAint predicts a probability score, which varies from 0-9 for each residue of protein sequence. At default 0.0 threshold, probability scores ranges between 0-4 and 5-9 predicted as non-interacting and interacting residues respectively.